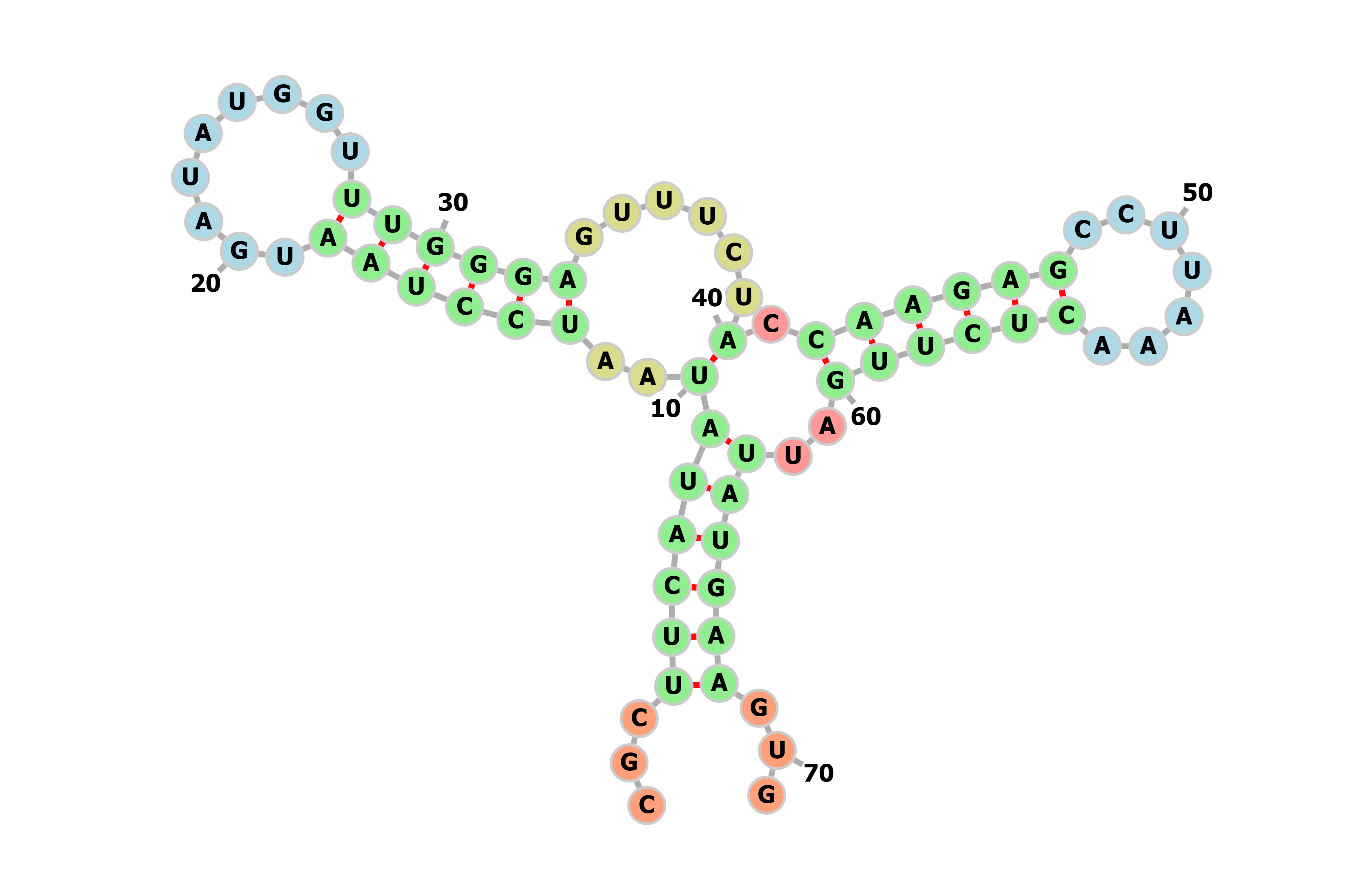
COMPRASION

| index | Standard value | Current value | estimate |
| CAI | 0.6~1 | 0.621-0.663 | normal |
| MFE | indefinite | -33.5 - -33.3 | normal |
| GC | indefinite | 2 | normal |
| TIME | 0~4h | 0 | slower |
Option1: Your mRNA design will start from here, input your ORF sequence, check the relevant parameters, click submit, and then wait for the optimized mRNA sequence to be generated.
If you only want to design ORF, just ignore Option2
Your mRNA design will start from here, input your ORF sequence, check the relevant parameters, click submit, and then wait for the optimized mRNA sequence to be generated.
If you only want to design ORF, just ignore Option2
 Your mRNA design will start from here, input your ORF sequence, check the relevant parameters, click submit, and then wait for the optimized mRNA sequence to be generated.
If you only want to design ORF, just ignore Option2
Your mRNA design will start from here, input your ORF sequence, check the relevant parameters, click submit, and then wait for the optimized mRNA sequence to be generated.
If you only want to design ORF, just ignore Option2
 Optiseed mRNA vaccine design
Optiseed mRNA vaccine design